
We finish the first half of the semester by studying how molecular models based on classical mechanics give rise to macroscopic phenomena. Students turn in their mid-term projects at the end of the week.
Simple molecular dynamics models are often constructed using the Lennard-Jones potential.
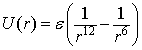
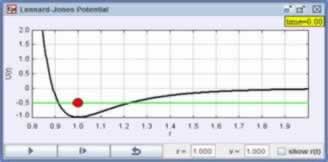
The EJS Lennard-Jones Potential Model shows the dynamics of a particle of mass m within this potential. You can drag particle to change its position and you can drag the energy-line to change its total energy.
The Lennard-Jones potential function was proposed by John Lennard-Jones as a reasonably accurate model of interactions between atoms because the dipole-dipole attraction has a 1/r6 potential energy. The 1/r12 is added to model the strong repulsion that occurs then the electron orbits overlap. The binding energy epsilon is the depth of the potential well and minimum molecular separation are set equal to unity.
The EJS Lennard-Jones Potential Model uses uses a natural system of units with the mass and the depth of the Lennard-Jones potential set equal to 1. For argon (for example), the unit of distance is 3.4 angstroms, the unit of mass is 40 atomic mass units, and the unit of energy is 0.01 electron-volts. The corresponding unit of time is then 2.2 picoseconds, the unit of velocity is 160 meters per second.
Users can drag the the red marker within the potential energy display to set the mechanical energy and the value of r. Study how this action is implemented in the Drag Energy custom method. An EJS action can limit the motion of a drag action by changing the value of variables that are connected to position and size properties.
Because the potential energy plot x and y axes are the radial separation r and the potential energy U(r), the format for the coordinate display properties have been changed in the plotting panel. These small improvements are easy to do and can make a model's user interfaced more attractive and easier to understand.
The following models use the Lennard-Jones potential to model molecular dynamics.
Additional models may be be posted for self-study.
The Lennard-Jones Potential Model was created for teaching computer modeling by Wolfgang Christian using the Easy Java Simulations (EJS) version 4.1 authoring and modeling tool. You can examine and modify a compiled EJS model if you run the model (double click on the model's jar file), right-click within a plot, and select "Open Ejs Model" from the pop-up menu. You must, of course, have EJS installed on your computer.
Information about Ejs is available at: <http://www.um.es/fem/Ejs/> and in the OSP comPADRE collection <http://www.compadre.org/OSP/>.